Cytologists have characterized numerous structural rearrangements in chromosomes, including partial duplications, deletions, inversions, and translocations. Duplications and deletions often produce offspring that survive but exhibit physical and mental abnormalities. Cri-du-chat (from the French for “cry of the cat”) is a syndrome associated with nervous system abnormalities and identifiable physical features that results from a deletion of most of the small arm of chromosome 5 (Figure 7.11). Infants with this genotype emit a characteristic high-pitched cry upon which the disorder’s name is based.

Chromosome inversions and translocations can be identified by observing cells during meiosis because homologous chromosomes with a rearrangement in one of the pair must contort to maintain appropriate gene alignment and pair effectively during prophase I.
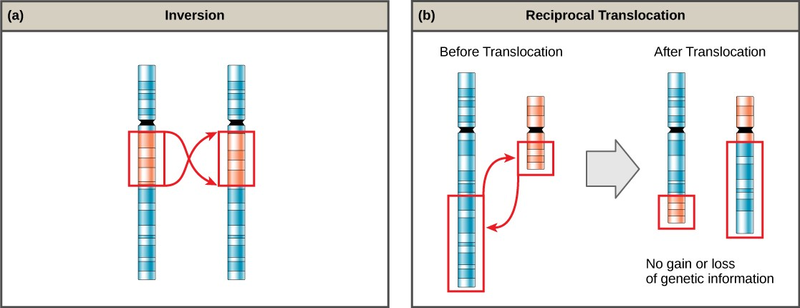
A chromosome inversion is the detachment, 180° rotation, and reinsertion of part of a chromosome (Figure 7.12). Unless they disrupt a gene sequence, inversions only change the orientation of genes and are likely to have more mild effects than aneuploid errors.
Evolution In Action
The Chromosome 18 Inversion
Not all structural rearrangements of chromosomes produce nonviable, impaired, or
infertile individuals. In rare instances, such a change can result in the evolution of a new species. In fact, an inversion in chromosome 18 appears to have contributed to the evolution of
humans. This inversion is not present in our closest genetic relatives, the chimpanzees.
The chromosome 18 inversion is believed to have occurred in early humans following their
divergence from a common ancestor with chimpanzees approximately five million years ago. Researchers have suggested that a long stretch of DNA was duplicated on chromosome 18 of an ancestor
to humans, but that during the duplication it was inverted (inserted into the chromosome in reverse orientation.
A comparison of human and chimpanzee genes in the region of this inversion indicates that
two genes—ROCK1and USP14—are farther apart on human chromosome 18 than they are on the corresponding chimpanzee chromosome.
This suggests that one of the inversion breakpoints occurred between these two genes. Interestingly, humans and chimpanzees express USP14at distinct levels in specific cell types, including cortical cells and fibroblasts. Perhaps the
chromosome 18 inversion in an ancestral human repositioned specific genes and reset their expression levels in a useful way. Because both ROCK1 and USP14code for enzymes, a change in their expression could alter cellular function. It is not known how
this inversion contributed to hominid evolution, but it appears to be a significant factor in the divergence of humans from other primates 1
A translocation occurs when a segment of a chromosome dissociates and reattaches to a different, nonhomologous chromosome. Translocations can be benign or have devastating effects, depending on how the positions of genes are altered with respect to regulatory sequences. Notably, specific translocations have been associated with several cancers and with schizophrenia. Reciprocal translocations result from the exchange of chromosome segments between two nonhomologous chromosomes such that there is no gain or loss of genetic information (Figure 7.12).
- 4460 reads






